Motivating Example
Motivating Example
Kick off the workshop by exploring a real data set using R!
Goal: get the flavor of using R for data management and exploration
Don’t worry about understanding all the coding right away
We will go back and explain how it all works in detail
Follow along using 2-MotivatingExample.R
Tip: Using “#” in your R File to take notes
- Super important to document your code!
Getting Started
Let’s begin by installing and loading tidyverse:
This workshop will focus largely on a group of packages that live together under the name tidyverse.
tidyverse includes the following well known packages:
ggplot2dplyrtidyrreadr- and more!
Salmonella Shedding Data
Fecal Salmonella shedding recorded over the course of 21 days
Several variables were recorded for each pig at certain time points:
- Amount of Salmonella in feces (log10)
- Pig weight (kg)
- Dietary treatment group
Primary Question: Do the different dietary treatments affect Salmonella shedding?
First look at data in R
Let’s use R to look at the top few rows of the Salmonella shedding data set. First, we load the data set using read_csv:
Now, we use the head function to look at the first 6 rows of the data:
# A tibble: 6 × 12
pignum time_point pig_weight temp pan_wt wet_wt Dry_wt_pan Dry_weight
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 77 2 NA 103. 1.02 1.09 1.31 0.29
2 87 2 NA 104. 1.04 1.02 1.33 0.29
3 122 2 NA 104. 1.02 0.99 1.11 0.09
4 160 2 NA 104. 1.09 1 1.36 0.27
5 191 2 NA 104. 1.03 1.02 1.27 0.24
6 224 2 NA 103 1.08 1 1.39 0.31
# ℹ 4 more variables: percent_drymatter <dbl>, daily_shedding <dbl>,
# treatment <chr>, total_shedding <dbl>Salmonella Shedding Data: Attributes
How big is this data set, and what types of variables are in each column?
spc_tbl_ [295 × 12] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
$ pignum : num [1:295] 77 87 122 160 191 224 337 345 419 458 ...
$ time_point : num [1:295] 2 2 2 2 2 2 2 2 2 2 ...
$ pig_weight : num [1:295] NA NA NA NA NA NA NA NA NA NA ...
$ temp : num [1:295] 103 104 104 104 104 ...
$ pan_wt : num [1:295] 1.02 1.04 1.02 1.09 1.03 1.08 1.03 1.09 1.07 1.04 ...
$ wet_wt : num [1:295] 1.09 1.02 0.99 1 1.02 1 1.02 0.99 1.07 1.06 ...
$ Dry_wt_pan : num [1:295] 1.31 1.33 1.11 1.36 1.27 1.39 1.3 1.24 1.38 1.26 ...
$ Dry_weight : num [1:295] 0.29 0.29 0.09 0.27 0.24 0.31 0.27 0.15 0.31 0.22 ...
$ percent_drymatter: num [1:295] 26.61 28.43 9.09 27 23.53 ...
$ daily_shedding : num [1:295] 5.3 5.7 13.22 6.75 5.86 ...
$ treatment : chr [1:295] "control" "control" "control" "control" ...
$ total_shedding : num [1:295] 2.3 2.48 5.74 2.93 2.54 ...
- attr(*, "spec")=
.. cols(
.. pignum = col_double(),
.. time_point = col_double(),
.. pig_weight = col_double(),
.. temp = col_double(),
.. pan_wt = col_double(),
.. wet_wt = col_double(),
.. Dry_wt_pan = col_double(),
.. Dry_weight = col_double(),
.. percent_drymatter = col_double(),
.. daily_shedding = col_double(),
.. treatment = col_character(),
.. total_shedding = col_double()
.. )
- attr(*, "problems")=<externalptr> Salmonella Shedding: Variables
Let’s get a summary of the values for each variable:
pignum time_point pig_weight temp pan_wt
Min. : 6.0 Min. : 0.0 Min. : 9.48 Min. :101.2 Min. :1.000
1st Qu.:119.0 1st Qu.: 2.0 1st Qu.:16.02 1st Qu.:102.4 1st Qu.:1.030
Median :224.0 Median : 7.0 Median :19.78 Median :102.9 Median :1.050
Mean :234.7 Mean : 8.8 Mean :21.01 Mean :102.9 Mean :1.054
3rd Qu.:361.0 3rd Qu.:14.0 3rd Qu.:25.29 3rd Qu.:103.4 3rd Qu.:1.070
Max. :472.0 Max. :21.0 Max. :36.30 Max. :105.2 Max. :1.120
NA's :59 NA's :118 NA's :122
wet_wt Dry_wt_pan Dry_weight percent_drymatter
Min. :0.700 Min. :1.110 Min. :0.090 Min. : 9.091
1st Qu.:1.000 1st Qu.:1.300 1st Qu.:0.250 1st Qu.:24.292
Median :1.020 Median :1.320 Median :0.280 Median :26.923
Mean :1.026 Mean :1.322 Mean :0.268 Mean :26.109
3rd Qu.:1.050 3rd Qu.:1.353 3rd Qu.:0.290 3rd Qu.:28.431
Max. :1.110 Max. :1.460 Max. :0.380 Max. :36.893
NA's :123 NA's :123 NA's :123 NA's :123
daily_shedding treatment total_shedding
Min. : 0.000 Length:295 Min. : 0.000
1st Qu.: 0.000 Class :character 1st Qu.: 1.699
Median : 3.912 Mode :character Median :13.904
Mean : 3.765 Mean :17.423
3rd Qu.: 5.521 3rd Qu.:30.893
Max. :13.218 Max. :66.207
Scatterplots
Let’s look at the relationship between time point and Salmonella shedding.
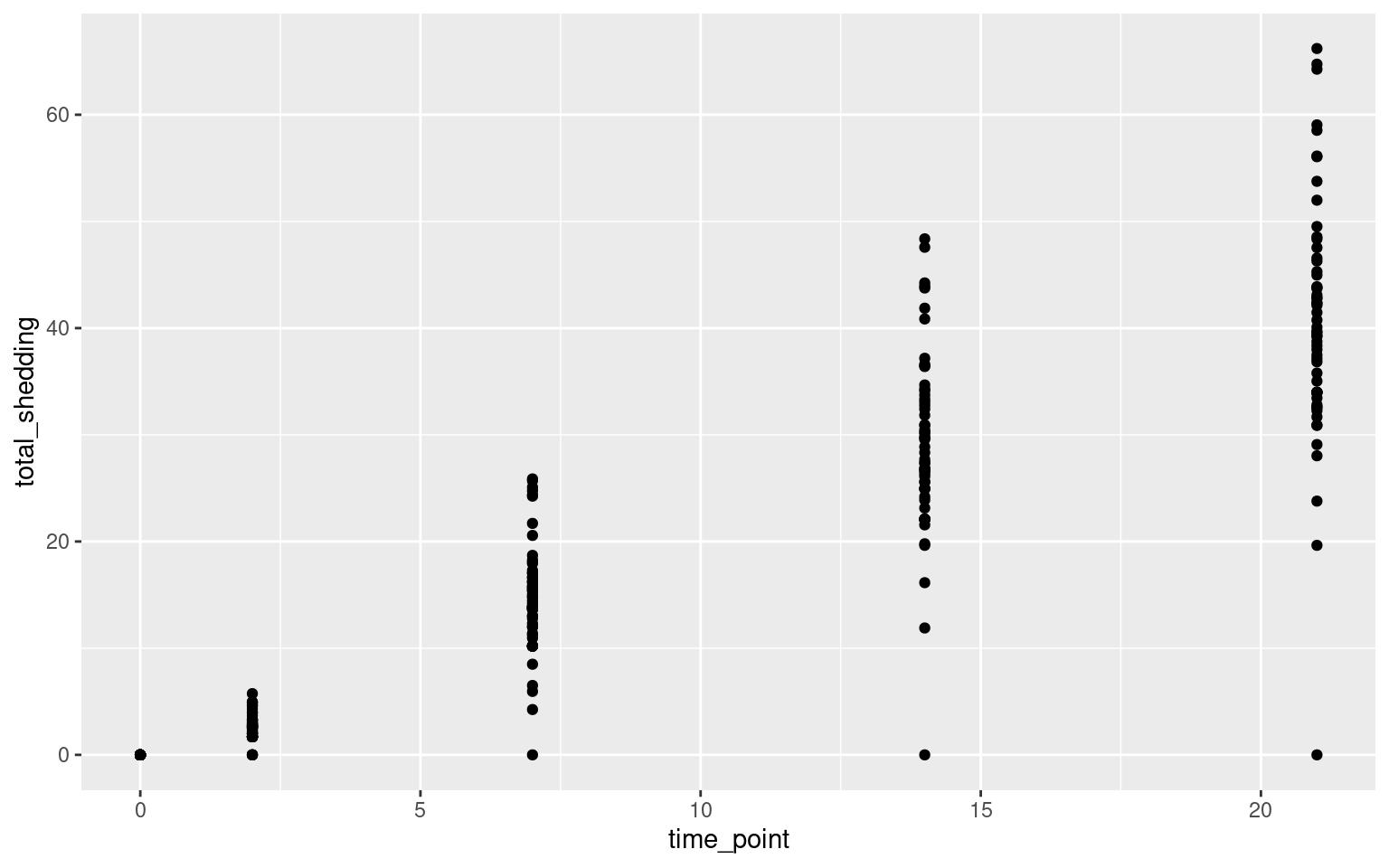
More Scatterplots
Color the points by treatment groups
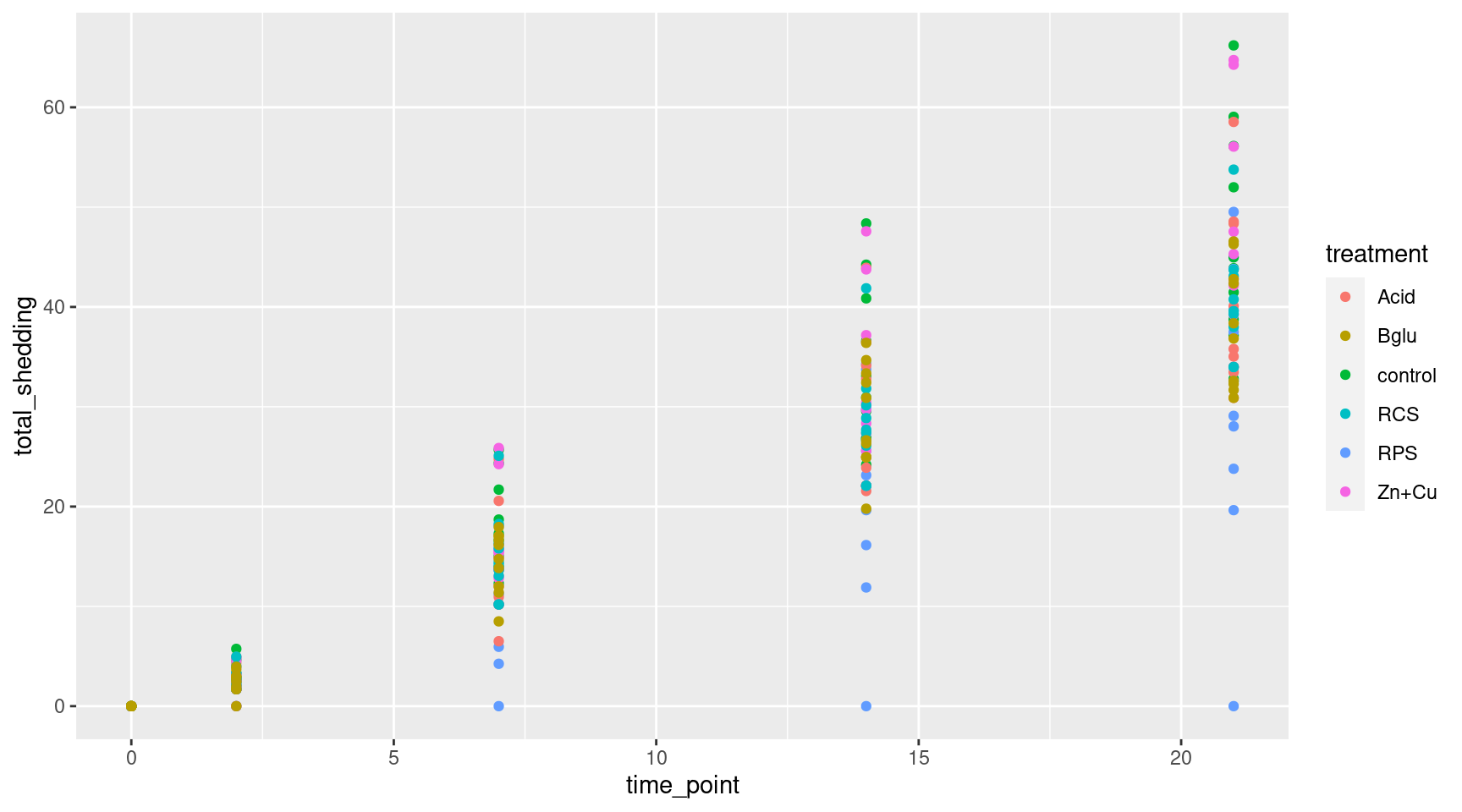
More Scatterplots
Switch to lines for easier reading
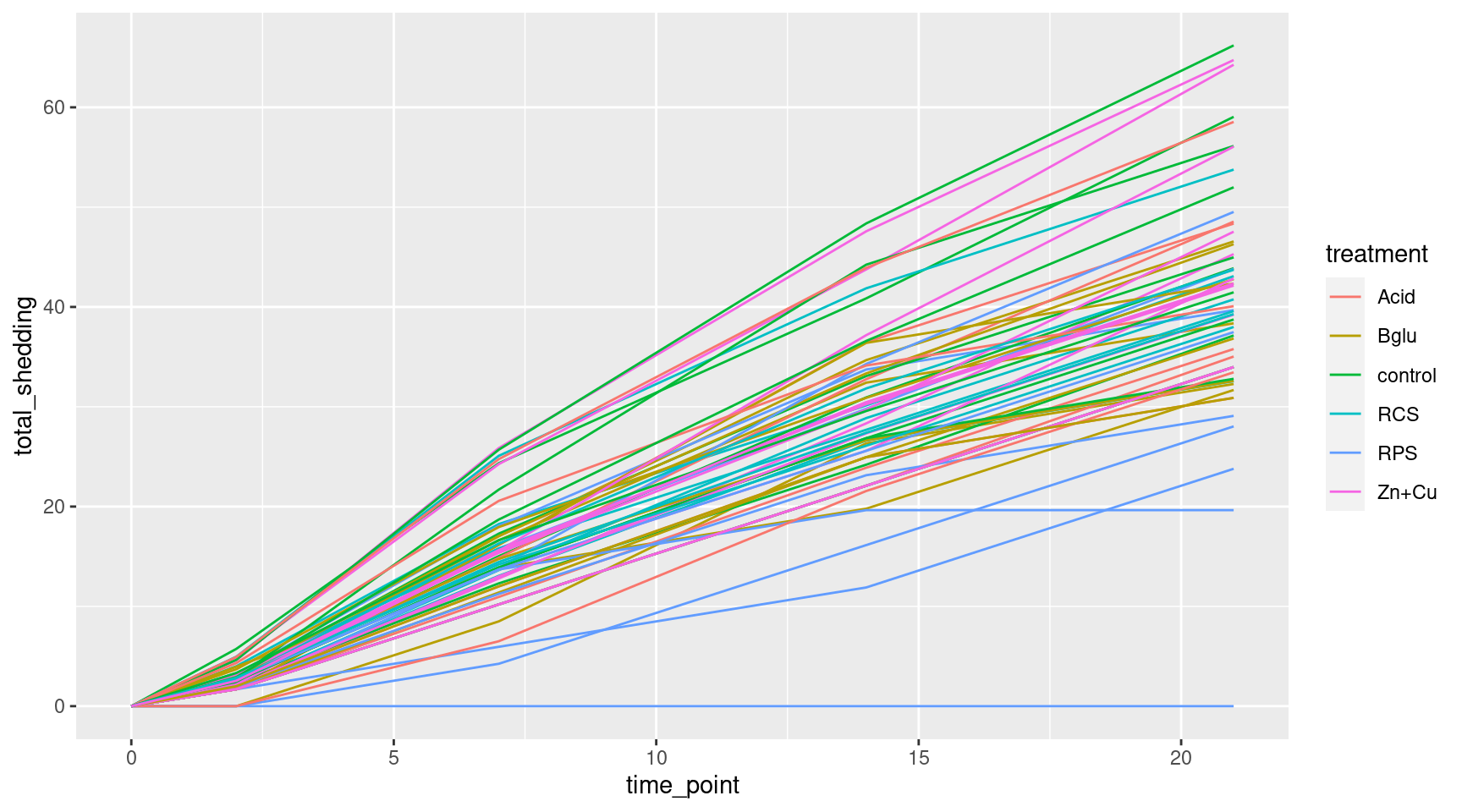
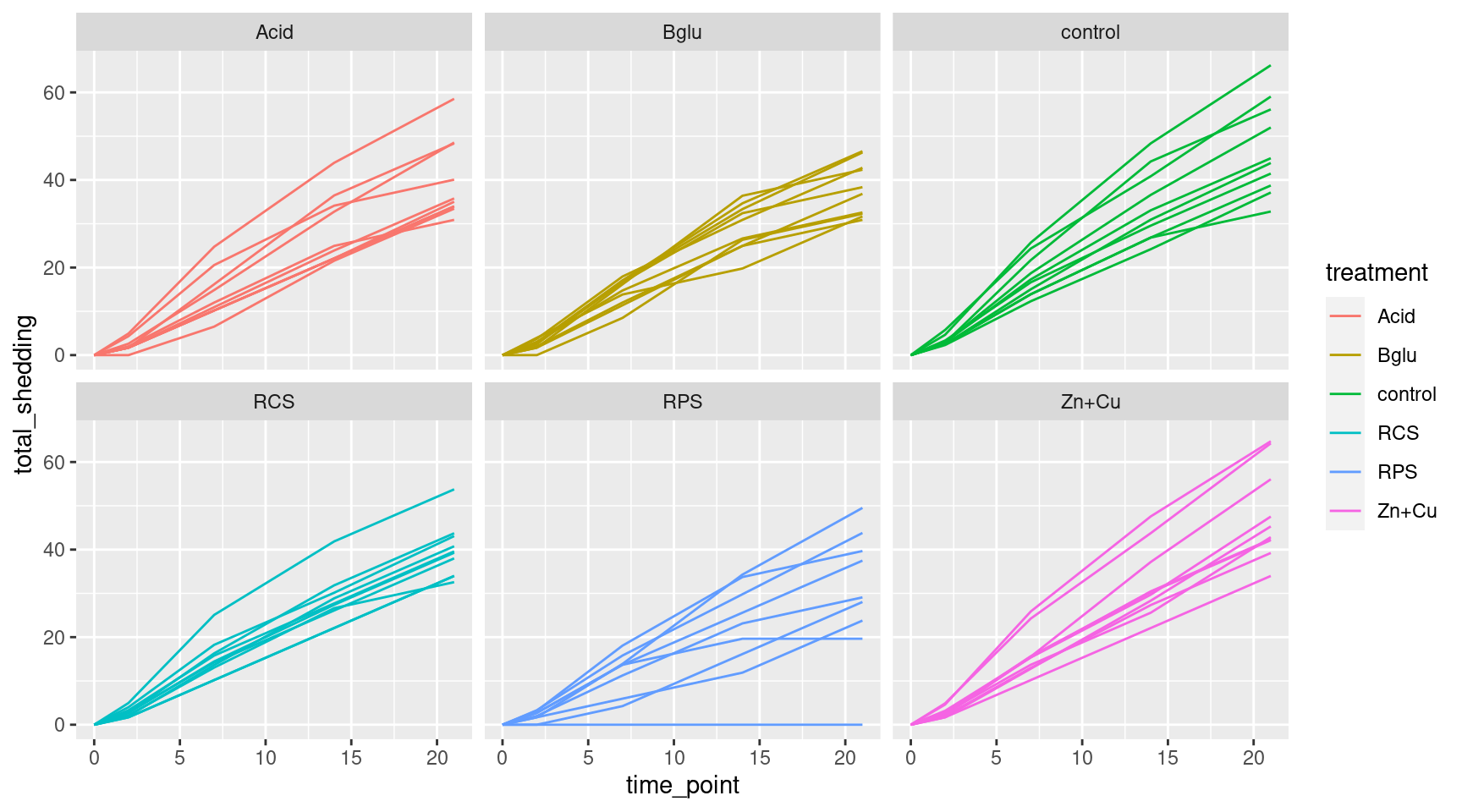
Even More Plots
Add faceting
Variable Creation and Data Filtering
We will make a new variable in the data: gain = weight at day 21 - weight at day 0.
We will then filter the data to only include the final values.
Histogram
Let’s look distribution of the final shedding values with a histogram
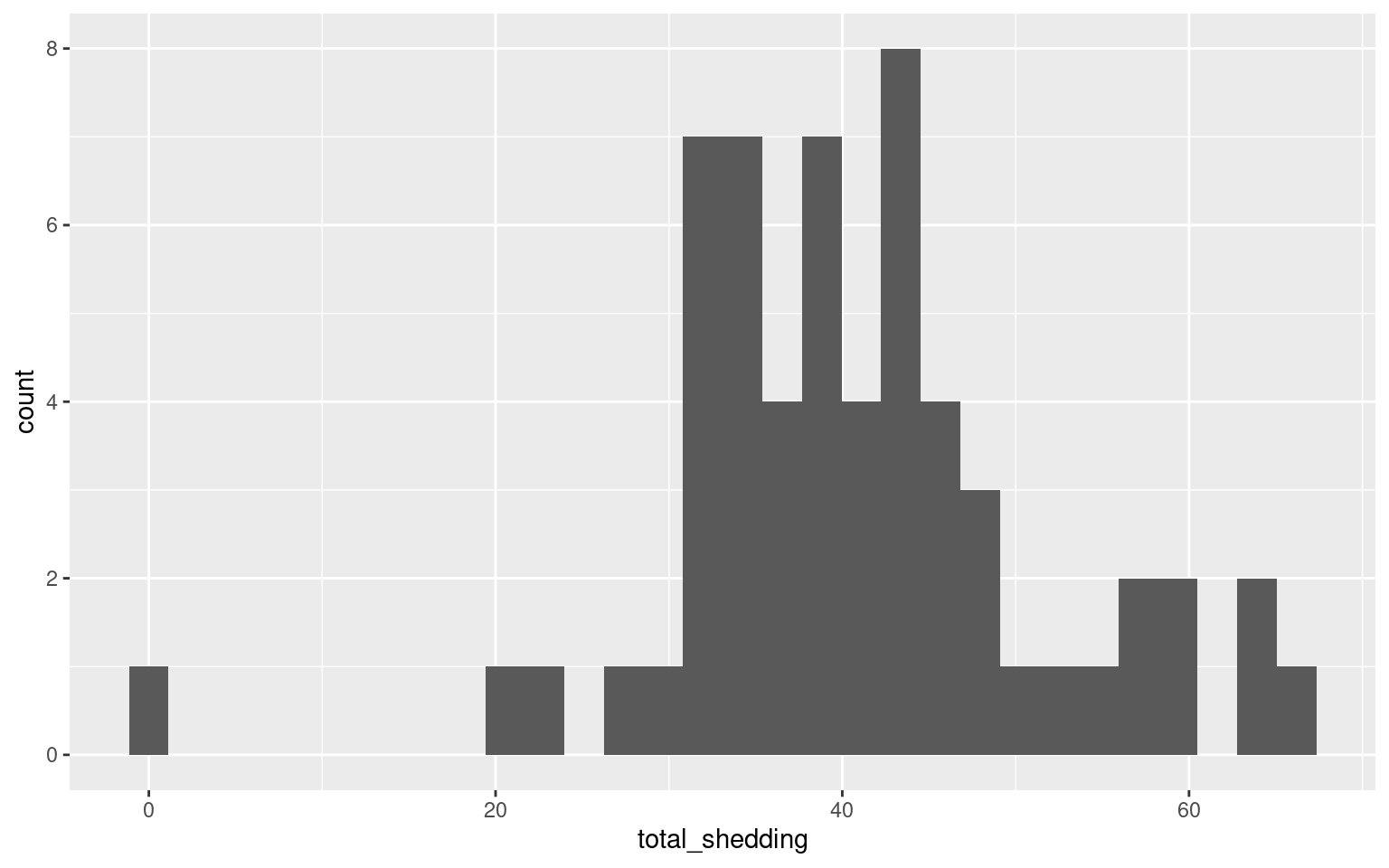
One pig did not shed at all…
Which pig, and what treatment did it receive?
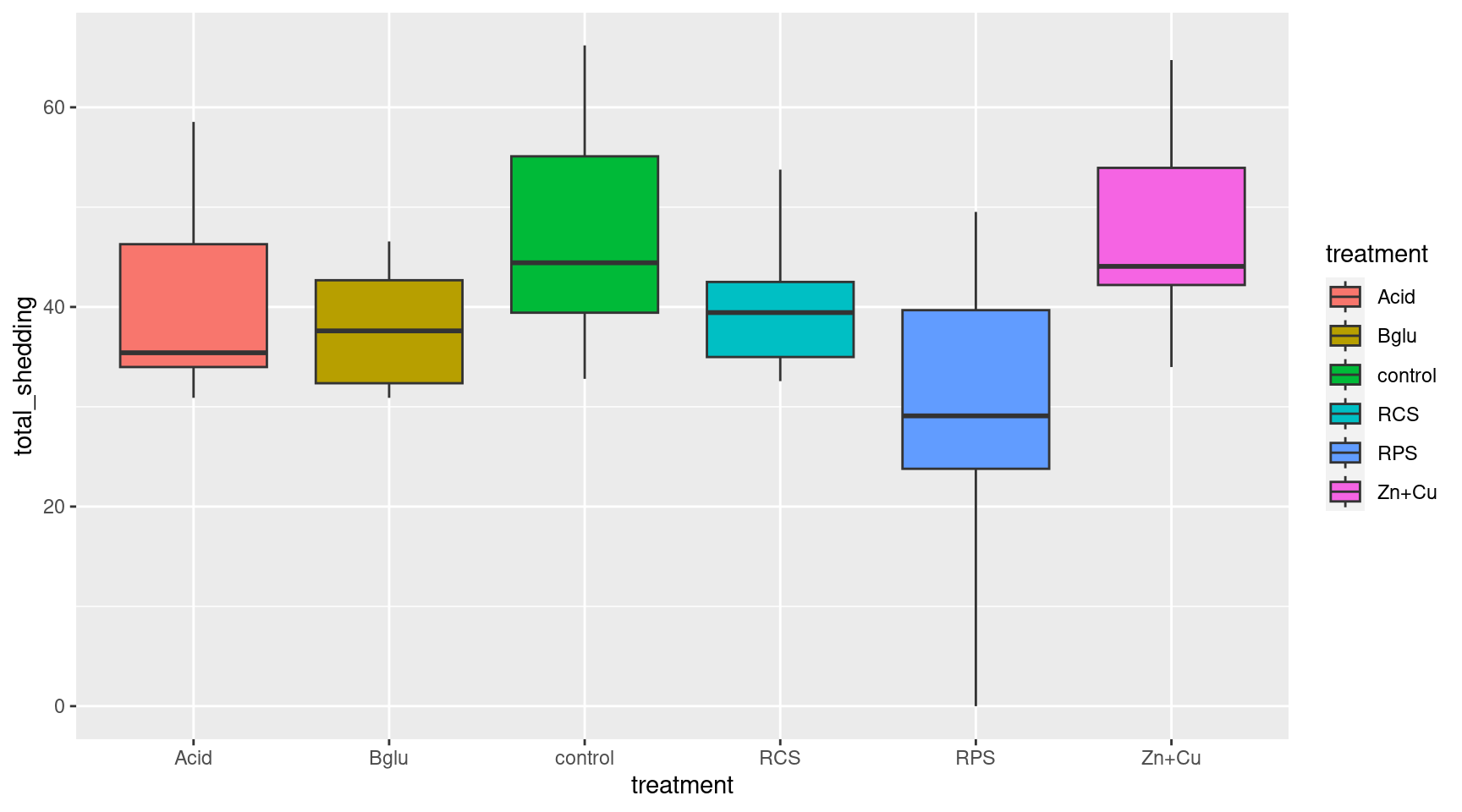
Shedding by Treatment
Look at the average cumulative shedding value for each treatment separately
Statistical Significance
There is a difference, but is it statistically significant?
Boxplots
We could compare the cumulative shedding values of each treatment group with a side by side boxplot
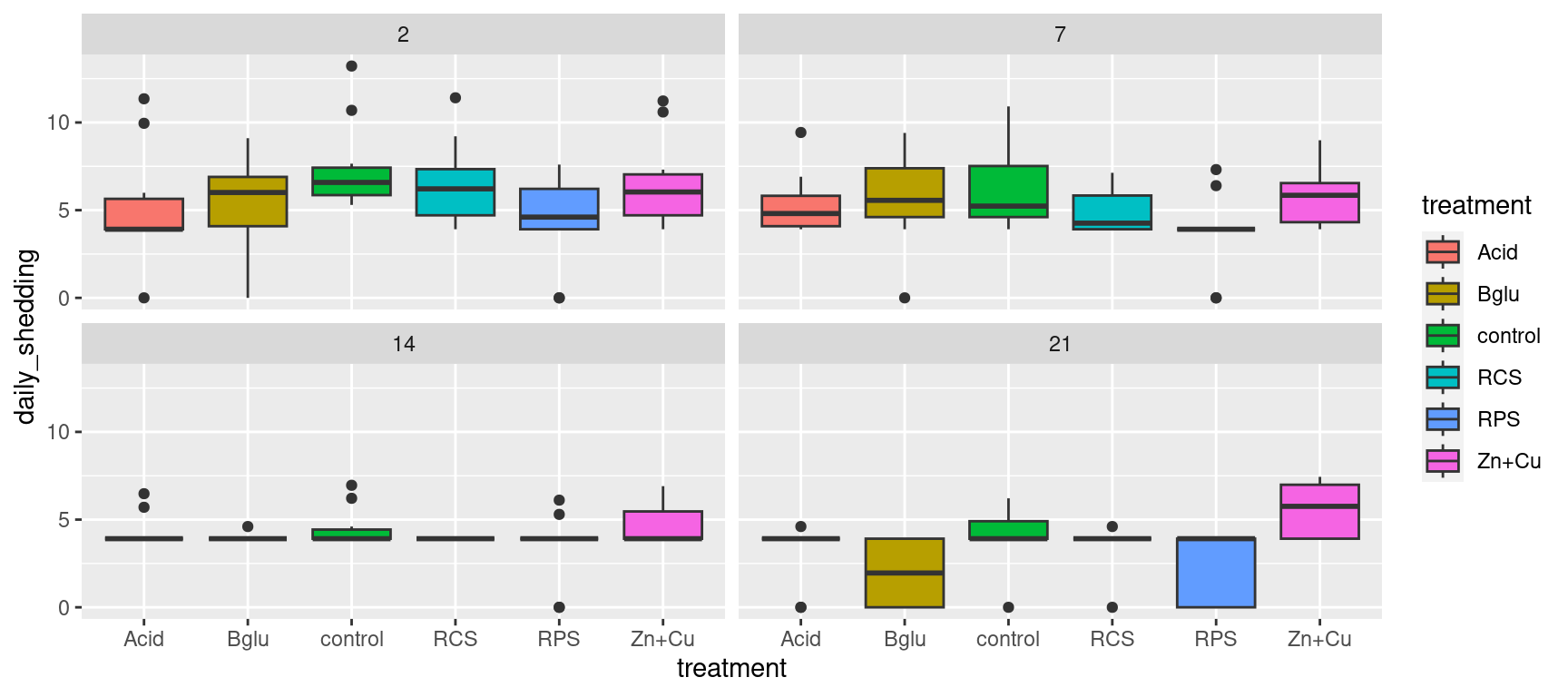
Boxplots Again
Alternatively, we could use the original data to compare the daily shedding values of each treatment group with side by side boxplots over time
Your Turn
Try playing with chunks of code from this session to further investigate the Salmonella shedding data:
Get a summary of the daily shedding values (use the
sheddata set)Make side by side boxplots of final weight gain by treatment group (use the
final_sheddata set)Compute a Wilcox test for control vs. the “Bglu” treatment group